| Gastroenterology Research, ISSN 1918-2805 print, 1918-2813 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, Gastroenterol Res and Elmer Press Inc |
| Journal website https://gr.elmerpub.com |
Original Article
Volume 18, Number 5, October 2025, pages 247-253
Enabling Drug-Induced Liver Injury Surveillance Through Automated Medication Extraction From Clinical Notes: A Medical Information Mart for Intensive Care IV Real-World Large Language Models Validation Study
Thanathip Suenghataiphorna, e, Kanachai Boonpiraksb, Vitchapong Prasitsumritc, Narathorn Kulthamrongsrid, Pojsakorn Danpanichkulc
aDepartment of Internal Medicine, Griffin Hospital, Derby, CT, USA
bUniversity of Kansas Medical Center, Kansas City, KS, USA
cDepartment of Internal Medicine, Texas Tech University Health Science Center, Lubbock, TX, USA
dUniversity of Hawaii, Honolulu, HI, USA
eCorresponding Author: Thanathip Suenghataiphorn, Department of Internal Medicine, Griffin Hospital, Derby, CT 06418, USA
Manuscript submitted July 10, 2025, accepted September 20, 2025, published online October 9, 2025
Short title: DILI Surveillance using LLM: MIMIC-IV Study
doi: https://doi.org/10.14740/gr2062
| Abstract | ▴Top |
Background: Drug-induced liver injury (DILI) presents a significant diagnostic challenge, often leading to delayed detection. Unstructured clinical notes contain comprehensive medication data vital for DILI surveillance but are difficult to analyze systematically. Large language models (LLMs) show promise for automated extraction but require real-world clinical data validation to assess feasibility for clinical applications like DILI surveillance.
Methods: We retrospectively validated an LLM system on 100 randomly sampled Medical Information Mart for Intensive Care IV (MIMIC-IV) discharge summaries. Gold standard unique medication lists were derived via manual annotation and manual deduplication based on normalized drug names. LLM outputs underwent identical deduplication. Performance was assessed using precision, recall, and F1-score comparing deduplicated lists. MIMIC-IV data use agreement (DUA) compliance was ensured.
Results: Comparison yielded a precision of 0.85, recall of 1.00, and an F1-score of 0.92 for unique medication identification. The 174 false positives resulted from parsing or normalization errors; no medication hallucinations occurred. A subsequent DILI database lookup failed for approximately 6.2% of correctly identified unique medications, evaluated as a separate feasibility measure.
Conclusions: The LLM demonstrates high accuracy and perfect recall for unique medication extraction and identification from complex clinical notes, establishing technical feasibility. This represents a feasible and possible integration of LLM towards developing automated tools for enhanced DILI surveillance and improved patient safety.
Keywords: Drug-induced liver injury; Artificial intelligence; Large language model; Machine learning
| Introduction | ▴Top |
Drug-induced liver injury (DILI) remains a significant clinical challenge for hepatologists worldwide, ranking as a leading cause of acute liver failure in Western countries and contributing substantially to patient morbidity, mortality, and healthcare costs [1]. Diagnosing DILI is complex, often relying on recognizing suggestive clinical patterns, temporal associations with drug initiation, and the exclusion of competing etiologies, a process inherently susceptible to delays [2]. Timely identification of potential DILI is paramount to mitigating the risk of progression to severe, potentially irreversible liver damage, highlighting an urgent need for improved surveillance methods [3].
Electronic health records (EHRs) offer a rich repository of longitudinal patient data, including medication exposures, which could theoretically facilitate earlier DILI detection [4]. However, current surveillance often relies heavily on analyzing structured EHR data (e.g., medication orders, lab results) [5], which may not capture the full complexity of medication exposure, including timing, adjustments during hospitalization, over-the-counter medications, or discharge prescriptions accurately documented only within unstructured clinical notes, as seen in other kinds of study [6]. These narrative notes, such as admission histories, progress reports, and discharge summaries, contain invaluable details but present a formidable challenge for traditional automated analysis due to their inherent linguistic variability and complexity [7]. Moreover, current drug detection systems mainly rely on structured medication input data, flagging each medication [8], in which other parameters in patient care such as symptoms, laboratories value and patient history may not be accounted for.
The advent of artificial intelligence (AI), particularly the development of sophisticated large language models (LLMs), presents a transformative opportunity to unlock the information embedded within unstructured clinical text [9]. LLMs have demonstrated remarkable capabilities in natural language understanding and generation, suggesting potential utility in tasks like extracting salient clinical information, including medication details, from narrative notes [10]. While preliminary studies have explored LLMs for various healthcare applications, including medication extraction in limited contexts, rigorous validation using complex [11], real-world clinical data is essential before considering their use in high-stakes applications like patient safety surveillance [12].
This study addresses this critical gap by evaluating the performance of an LLM-based system specifically designed for medication extraction when applied to a diverse set of real-world, unstructured discharge summaries from the large MIMIC-IV critical care database. Accurate and comprehensive medication list generation is a prerequisite for any subsequent automated DILI risk assessment system. Therefore, the primary objective of this research was to rigorously quantify the accuracy of a possible LLM system in identifying the complete, unique list of medications documented throughout the continuum of care captured in discharge summaries, thereby assessing its feasibility as a foundational component for future automated DILI surveillance tools.
| Materials and Methods | ▴Top |
Study design
This study utilized a retrospective validation design to evaluate the performance of an LLM-based system developed for automated medication extraction. The accuracy of the system was assessed by comparing its output against a manually curated ground truth derived from unstructured clinical discharge summaries obtained from a large, publicly available critical care database.
Data source
The data for this validation study were sourced from the Medical Information Mart for Intensive Care IV (MIMIC-IV) database (version 2.2). MIMIC-IV is a large, deidentified, publicly accessible database comprising comprehensive clinical data related to patients admitted to critical care units at the Beth Israel Deaconess Medical Center (Boston, MA, USA) between 2008 and 2019. For this analysis, we utilized the unstructured clinical text from discharge summary notes contained within the MIMIC-IV dataset. A cohort of 100 discharge summaries was selected from this dataset via simple random sampling to serve as the basis for manual annotation and system performance evaluation. Table 1 shows the data information.
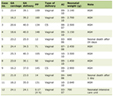 Click to view | Table 1. Descriptive Statistics for the Data Cohort |
Ethical considerations
This study utilized the deidentified MIMIC-IV database (version 2.2), accessed under a PhysioNet restricted data use agreement (DUA). The project adhered strictly to the data privacy and security stipulations outlined in the DUA. For this study, Institutional Review Board (IRB) approval and informed consent were not applicable.
LLM system architecture
Figure 1 shows the system architecture. The LLM system evaluated in this study processes unstructured clinical text to extract key medication information. The core components include an input layer for handling raw discharge summary text, an LLM model utilizing Llama-4-Scout-17B-16E-Instruct with engineered prompts requesting medication name, normalized name, dosage, frequency, and date in a structured JSON format, and an output layer generating the structured list. We selected the Llama 4 family as it provides the most efficient model of providing the required information. A downstream DILI linkage component normalizes extracted medication names and employs fuzzy against a combined DILI database compiled from DILIrank [13] and LiverTox [14] sources. The TRIPOD-LLM checklist is provided (Supplementary Material 1, gr.elmerpub.com). The system prompt used is shown (Supplementary Material 2, gr.elmerpub.com), and the programming information, accessible online, is presented (Supplementary Material 3, gr.elmerpub.com).
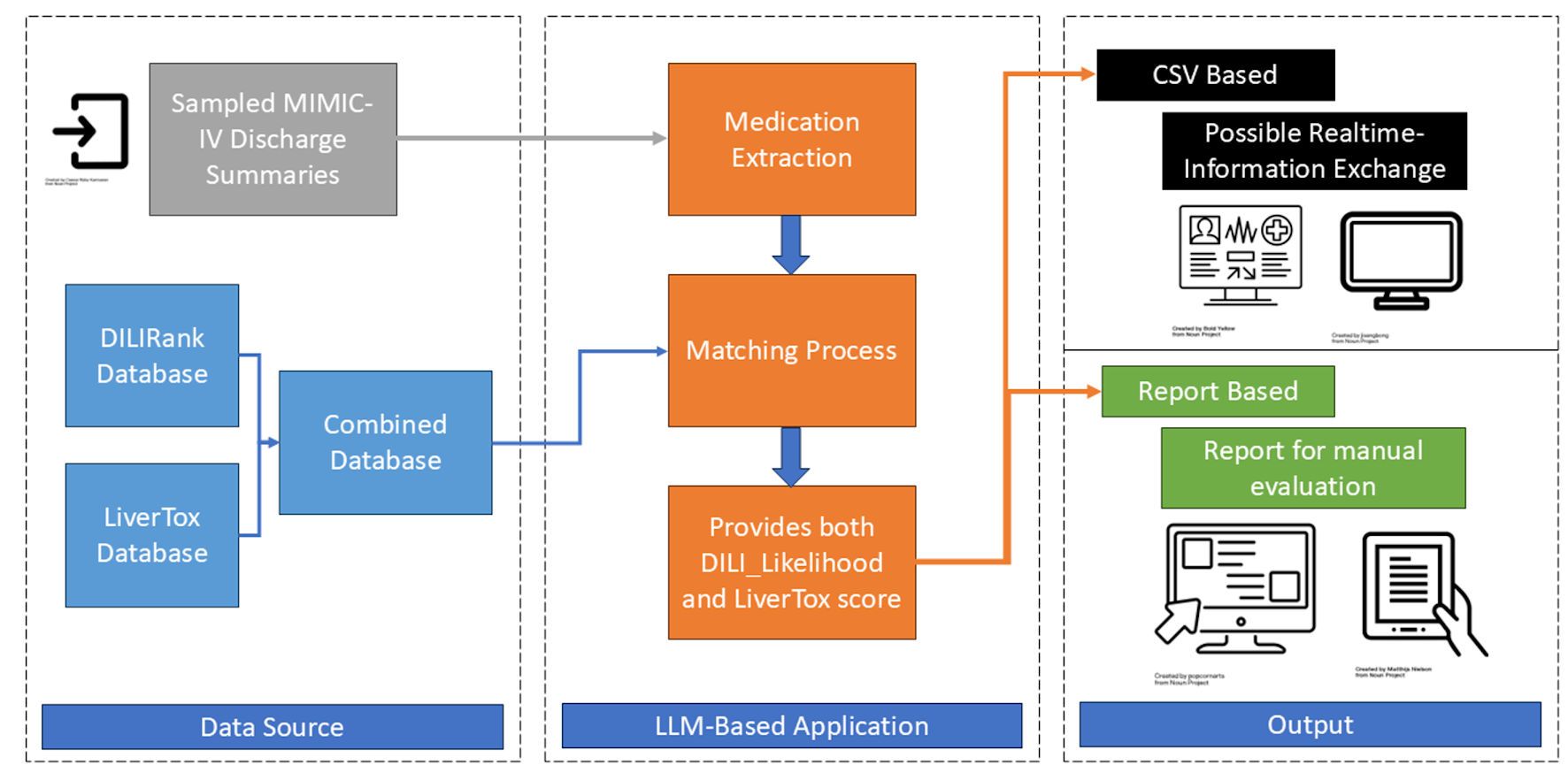 Click for large image | Figure 1. System architect. MIMIC-IV: Medical Information Mart for Intensive Care IV; DILI: drug-induced liver injury; LLM: large language models. |
Medication extraction scope
The analysis included the entire text of the discharge summary, aiming to identify medications mentioned in relation to admission, hospital course, and discharge periods to capture comprehensive drug exposure relevant to DILI.
Validation methodology
Ground truth generation
Two independent reviewers (KB, VP) blinded to the LLM output manually annotated each of the 100 discharge summaries, identifying all medication mentions. A third independent reviewer (PD) resolved discrepancies between the initial reviewers to establish a final “gold standard” list of medication mentions for each note.
Deduplication process
Both the adjudicated ground truth list and the raw LLM output list for each note underwent a manual deduplication process based on the normalized medication name, resulting in a list of unique medications per note for both ground truth and LLM output.
Performance metrics definition
Performance was evaluated by comparing the deduplicated LLM list against the deduplicated ground truth list for each note. A true positive (TP) was defined as a unique normalized medication name present in both lists. A false positive (FP) was defined as a unique normalized medication name present in the LLM list but not the ground truth list. A false negative (FN) was defined as a unique normalized medication name present in the ground truth list but not the LLM list.
To explicitly confirm the absence of FNs, the third adjudicating reviewer performed a final check, systematically comparing the final deduplicated ground truth list against the deduplicated LLM list for each of the 100 notes.
Statistical analysis
Counts for TP, FP, and FN were aggregated across all 100 notes. Overall precision, recall, and F1-score were calculated using standard formulas (precision = TP/(TP + FP); recall = TP/(TP + FN); F1 = 2 × (precision × recall)/(precision + recall)). The DILI lookup failure rate was calculated separately as the number of correctly identified unique medications for which the lookup failed, divided by the total number of unique medications correctly identified by the LLM extraction component.
| Results | ▴Top |
Medication extraction performance (deduplicated)
The overall study workflow, from sample selection to final analysis, is depicted in Figure 2. The LLM system was evaluated on 100 randomly selected MIMIC-IV discharge summaries. After processing these notes and performing manual deduplication based on normalized medication names, the system generated a total of 1,236 unique medication extractions.
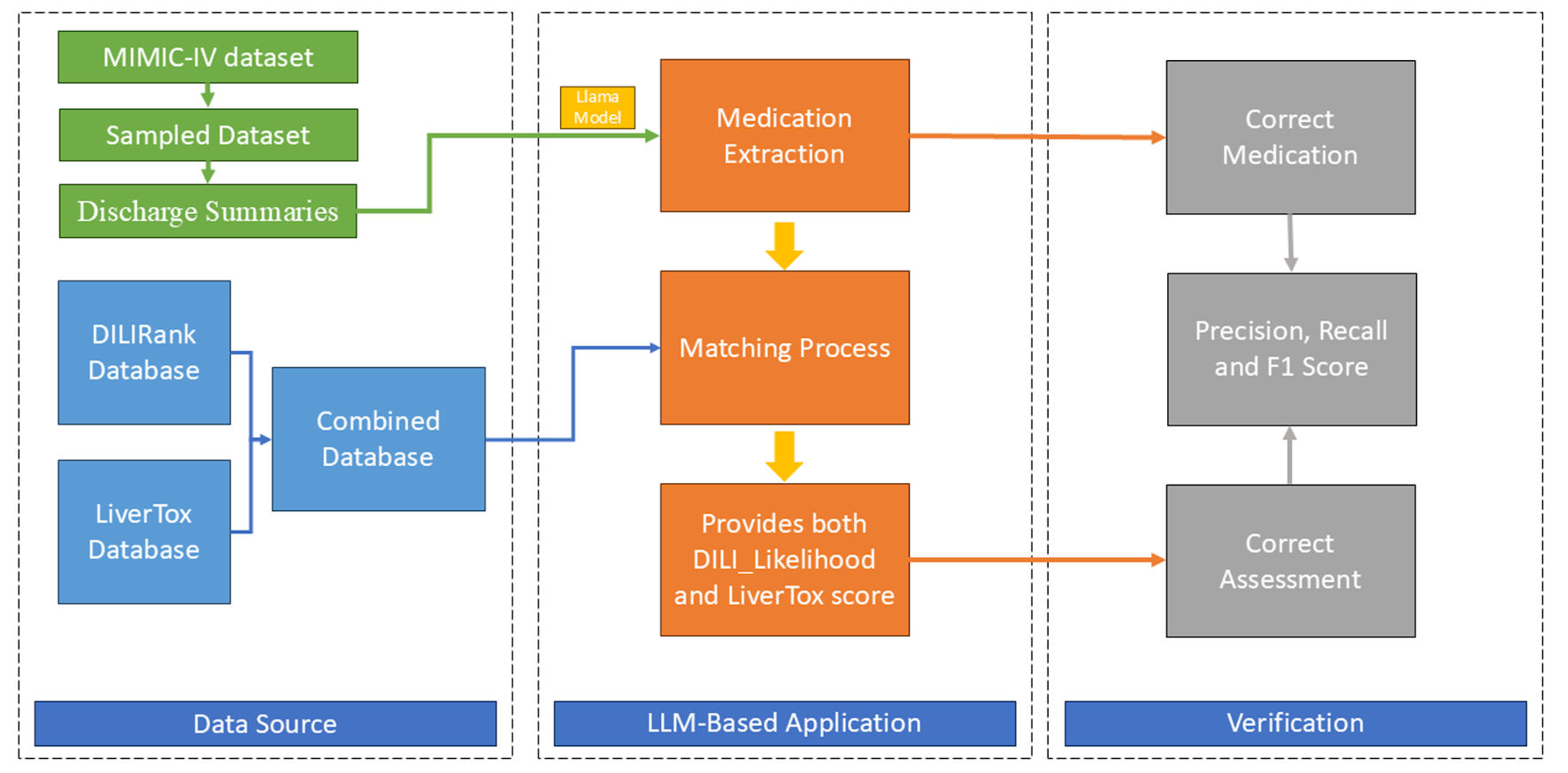 Click for large image | Figure 2. Study flowchart. MIMIC-IV: Medical Information Mart for Intensive Care IV; DILI: drug-induced liver injury; LLM: large language models. |
Comparison of the deduplicated LLM output against the deduplicated ground truth list yielded 996 TPs, 174 FPs, and 0 FNs across the 100 notes. Based on these aggregated counts, the LLM system achieved an overall precision of 0.85, a recall of 1.00, and an F1-score of 0.92 for the task of unique medication extraction (Table 2). The perfect recall (1.00) signifies that the system successfully identified all unique medications present in the manually adjudicated ground truth lists for the evaluated notes.
 Click to view | Table 2. Performance Metrics for Dataset |
Error analysis (FPs)
An analysis of the 174 FPs revealed that all instances were attributable to errors in the parsing or normalization of medication names that were correctly identified by the LLM as being present in the text. Common error types included incomplete extraction of compound drug names (e.g., extracting only “amoxicillin” when “amoxicillin-clavulanate” was documented) or failure to normalize variations in drug nomenclature to the required standard format. Importantly, no instances of hallucination (i.e., extraction of medications not mentioned in the source text) were observed among the deduplicated results.
DILI linkage outcome
As a secondary assessment relevant to the system’s potential application in DILI surveillance, we evaluated the success rate of the downstream DILI database linkage component. Of the 1,062 unique medications correctly identified by the LLM (996 TP + 66 errors where DILI lookup failed but extraction was correct), the fuzzy matching algorithm successfully linked 996 (TPs) to an entry in the combined DILI database. The remaining 66 correctly identified unique medications (approximately 6.2%) could not be matched to a database entry using the predefined threshold, representing failures in the DILI linkage step rather than the initial LLM extraction.
| Discussion | ▴Top |
This study evaluated the real-world performance of an LLM-based system for extracting comprehensive, unique medication lists from complex clinical discharge summaries, a critical prerequisite for developing automated DILI surveillance tools. Our principal finding is that the system demonstrated substantial accuracy, achieving an F1-score of 0.92, underpinned by a perfect recall (1.00) and a precision of 0.85 when evaluated on deduplicated medication lists derived from 100 MIMIC-IV discharge summaries. This indicates that, within our validated sample, the LLM successfully identified every unique medication documented in the reference standard established by rigorous manual review.
In the context of DILI surveillance, missing even a single potentially offending drug could have significant clinical consequences, especially when mapping multiple drugs to assess causality [15]. The ability of the LLM to capture the complete set of unique medications mentioned throughout the discharge summary - spanning admission, hospital course, and discharge periods - suggests its potential utility in constructing accurate patient medication profiles necessary for risk assessment. While previous studies have employed various natural language processing (NLP) techniques for medication extraction [16, 17], achieving perfect recall on diverse, real-world clinical narratives remains challenging. Our findings highlight the capability of modern LLMs to handle the linguistic variability inherent in clinical text for comprehensive entity recognition.
While previous studies have employed various NLP techniques for medication extraction [18], achieving perfect recall on diverse, real-world clinical narratives remains a significant challenge for traditional rule-based or machine learning models that can struggle with linguistic variability and complex sentence structures. Our findings highlight the capability of modern LLMs to overcome some of these historical barriers to achieve comprehensive entity recognition. The precision of 0.85 indicates that while the system identified all relevant unique medications, it also generated some outputs (174 instances) that did not perfectly match the normalized ground truth format. Crucially, our error analysis revealed that these FPs were exclusively due to errors in parsing or normalization (e.g., handling complex drug names) rather than model hallucination or extracting non-medicinal terms. This suggests the LLM effectively grounds its extractions in the source text but may require further refinement or post-processing steps to consistently adhere to strict normalization standards, particularly for non-standard nomenclature or compound medications frequently encountered in clinical practice.
While the core medication extraction demonstrated high performance, the evaluation of the downstream DILI linkage component revealed challenges. Approximately 6.2% of correctly identified unique medications could not be matched to entries in our database. This highlights a separate bottleneck in developing a fully automated system. Potential reasons include the inherent limitations of existing curated DILI databases [19], which may not encompass all medications or formulations, and the challenges matching faces with highly variable or complex drug names not easily resolved to a canonical form present in the database. From a clinical perspective, a 6.2% linkage failure rate could lead to an underestimation of a patient’s true DILI risk if the unlinked drug is a known but rare hepatotoxin, or if a new drug’s potential for DILI is not yet cataloged. This could diminish the sensitivity of an automated surveillance system and potentially delay the identification of DILI signals, thereby impacting the tool’s clinical utility and trustworthiness. Improving drug name normalization and potentially integrating more comprehensive pharmacological knowledge bases will be essential for enhancing the reliability of automated DILI risk assignment [20].
This study has several strengths, including the use of a large, real-world clinical dataset (MIMIC-IV) representing complex patient cases compared to prior studies [21], a rigorous ground truth annotation process involving three independent reviewers, and the evaluation focused on clinically relevant unique medication lists. However, we acknowledge several limitations. First, while MIMIC-IV is diverse, it originates from a single academic medical center, potentially limiting generalizability. This setting may have uniform documentation practices and a specific formulary, potentially limiting the model’s generalizability. Real-world implementation across different institutions and healthcare systems would expose the model to significant variations in EHR platforms, note templates, colloquial language, and regional prescribing habits. Overcoming these challenges will require robust multi-center validation and potentially fine-tuning the model on more diverse datasets to ensure its performance is not biased by the idiosyncrasies of a single institution. Second, the system relies on a specific LLM accessed via a third-party application programming interface (API), raising considerations about potential model drift, cost, data privacy, and API reliance in broader implementations. Exploring the use of open-source models or fine-tuning smaller, domain-specific models could mitigate these challenges and offer a more sustainable path for clinical deployment. Third, our DILI linkage evaluation was preliminary; further work is needed to optimize and validate this component, especially concerns with bias [22]. Finally, this study focused on the technical accuracy of medication extraction; future prospective studies must evaluate the clinical utility of an end-to-end system in identifying actual DILI events and, crucially, assess its impact on clinician workflows and patient outcomes.
Despite these limitations, the promising results pave the way for important future research directions. Refining the normalization and parsing capabilities, potentially through targeted fine-tuning or improved prompt engineering, could further enhance precision. Developing more robust methods for linking extracted medications to comprehensive, regularly updated DILI knowledge sources is critical. Refining the normalization and parsing capabilities, potentially through targeted fine-tuning or the addition of a dedicated NLP post-processing module to specifically handle compound names and synonyms, could further enhance precision. Ultimately, real-time electronic medical record (EMR) monitoring - where the system could flag high-risk medication exposures in conjunction with relevant laboratory data (e.g., liver function tests (LFTs)) to alert clinicians to potential DILI risk proactively - will provide an additional tool for the physician to assess DILI risks. Exploring agentic AI frameworks could enable more autonomous monitoring and alerting within defined safety parameters.
Conclusions
In conclusion, this study demonstrates the substantial capability of an LLM system to accurately extract comprehensive, unique medication lists from complex, real-world clinical discharge summaries, achieving high precision and perfect recall in a rigorously validated cohort. While challenges remain in optimizing downstream linkage to DILI knowledge bases, the high fidelity of the core medication extraction establishes the technical feasibility of using LLMs for this critical task. This work provides a vital foundation for developing and integrating advanced AI tools into clinical practice to enhance drug safety surveillance and enable more timely detection of potential DILI.
Learning points
This study demonstrates that an advanced AI system can accurately read complex hospital discharge notes and identify all the unique medications a patient was taking. While the system sometimes made minor errors in how it formatted drug names, it never missed a medication or invented one that was not there. This shows that AI is a promising tool for automatically gathering complete medication information from patient records, which is a vital step towards building better systems to monitor for harmful drug side effects like liver injury and ultimately improve patient safety.
| Supplementary Material | ▴Top |
Suppl 1. TRIPOD-LLM checklist.
Suppl 2. The system prompt.
Suppl 3. The programming information.
Acknowledgments
None to declare.
Financial Disclosure
No funding was sought for this study.
Conflict of Interest
We declared no conflict of interest.
Informed Consent
Not applicable.
Author Contributions
Conceptualization: TSu, PD; Data curation: Tsu; Formal analysis: Tsu; Data source preparation: TSu, PD; Methodology/programming: Tsu; Validation: KB, VP; Writing - original draft: TSu, NK; Manuscript finalization: TSu, PD. All authors have read and approved the final version of the manuscript for submission.
Data Availability
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
| References | ▴Top |
- Hosack T, Damry D, Biswas S. Drug-induced liver injury: a comprehensive review. Therap Adv Gastroenterol. 2023;16:17562848231163410.
doi pubmed - Rockey DC, Seeff LB, Rochon J, Freston J, Chalasani N, Bonacini M, Fontana RJ, et al. Causality assessment in drug-induced liver injury using a structured expert opinion process: comparison to the Roussel-Uclaf causality assessment method. Hepatology. 2010;51(6):2117-2126.
doi pubmed - Stine JG, Lewis JH. Current and future directions in the treatment and prevention of drug-induced liver injury: a systematic review. Expert Rev Gastroenterol Hepatol. 2016;10(4):517-536.
doi pubmed - Huguet N, Kaufmann J, O'Malley J, Angier H, Hoopes M, DeVoe JE, Marino M. Using electronic health records in longitudinal studies: estimating patient attrition. Med Care. 2020;58(Suppl 6 1):S46-S52.
doi pubmed - Holmes JH, Beinlich J, Boland MR, Bowles KH, Chen Y, Cook TS, Demiris G, et al. Why is the electronic health record so challenging for research and clinical care? Methods Inf Med. 2021;60(1-02):32-48.
doi pubmed - Ruckdeschel JC, Riley M, Parsatharathy S, Chamarthi R, Rajagopal C, Hsu HS, Mangold D, et al. Unstructured data are superior to structured data for eliciting quantitative smoking history from the electronic health record. JCO Clin Cancer Inform. 2023;7:e2200155.
doi pubmed - Sedlakova J, Daniore P, Horn Wintsch A, Wolf M, Stanikic M, Haag C, Sieber C, et al. Challenges and best practices for digital unstructured data enrichment in health research: A systematic narrative review. PLOS Digit Health. 2023;2(10):e0000347.
doi pubmed - Liu S, Kawamoto K, Del Fiol G, Weir C, Malone DC, Reese TJ, Morgan K, et al. The potential for leveraging machine learning to filter medication alerts. J Am Med Inform Assoc. 2022;29(5):891-899.
doi pubmed - Meng X, Yan X, Zhang K, Liu D, Cui X, Yang Y, Zhang M, et al. The application of large language models in medicine: A scoping review. iScience. 2024;27(5):109713.
doi pubmed - Van Veen D, Van Uden C, Blankemeier L, Delbrouck JB, Aali A, Bluethgen C, Pareek A, et al. Adapted large language models can outperform medical experts in clinical text summarization. Nat Med. 2024;30(4):1134-1142.
doi pubmed - Lin C, Kuo CF. Roles and potential of large language models in healthcare: a comprehensive review. Biomed J. 2025;48(5):100868.
doi pubmed - Dennstadt F, Hastings J, Putora PM, Schmerder M, Cihoric N. Implementing large language models in healthcare while balancing control, collaboration, costs and security. NPJ Digit Med. 2025;8(1):143.
doi pubmed - Chen M, Suzuki A, Thakkar S, Yu K, Hu C, Tong W. DILIrank: the largest reference drug list ranked by the risk for developing drug-induced liver injury in humans. Drug Discov Today. 2016;21(4):648-653.
doi pubmed - Hoofnagle JH. In: Chapter 40 - LiverTox: a website on drug-induced liver injury. Kaplowitz N, DeLeve LD, Academic Press, Boston. 2013. p. 725-732.
- Suk KT, Kim DJ. Drug-induced liver injury: present and future. Clin Mol Hepatol. 2012;18(3):249-257.
doi pubmed - Chen L, Gu Y, Ji X, Sun Z, Li H, Gao Y, Huang Y. Extracting medications and associated adverse drug events using a natural language processing system combining knowledge base and deep learning. J Am Med Inform Assoc. 2020;27(1):56-64.
doi pubmed - Sezgin E, Hussain SA, Rust S, Huang Y. Extracting medical information from free-text and unstructured patient-generated health data using natural language processing methods: feasibility study with real-world data. JMIR Form Res. 2023;7:e43014.
doi pubmed - Rathee S, MacMahon M, Liu A, Katritsis NM, Youssef G, Hwang W, Wollman L, et al. DILI (C): an AI-based classifier to search for drug-induced liver injury literature. Front Genet. 2022;13:867946.
doi pubmed - Luo G, Shen Y, Yang L, Lu A, Xiang Z. A review of drug-induced liver injury databases. Arch Toxicol. 2017;91(9):3039-3049.
doi pubmed - Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011;18(4):441-448.
doi pubmed - Suenghataiphorn T, Danpanichkul P, Tribuddharat N, Kulthamrongsri N. Toward real-time detection of drug-induced liver injury using large language models: a feasibility study from clinical notes. J Clin Exp Hepatol. 2025;15(6):102627.
doi pubmed - Suenghataiphorn T, Tribuddharat N, Danpanichkul P, Kulthamrongsri N. Bias in large language models across clinical applications: A systematic review. arXiv. [csCL] 2025.
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Gastroenterology Research is published by Elmer Press Inc.